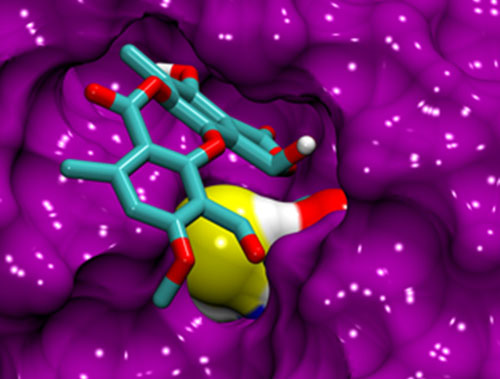
Suits you. This simulation of the p53 protein shows stictic acid fitted into the protein’s “reactivation pocket”. (Courtesy: Özlem Demir)
By Susan Curtis in Baltimore, US
At the 59th annual meeting of the Biophysical Society today, Rommie Amaro of the University of California, San Diego, highlighted the power of computational methods to speed up the discovery of new drugs to treat diseases as diverse as flu and cancer. Amaro focused on a recent project conducted while she was at the University of California, Irvine, to identify compounds that could play a vital role in future anti-cancer drugs by helping to reactive a molecule called p53 that is known to inhibit the formation of cancer cells.
“Using our computational approach, we have discovered more candidate compounds in the last 12 months than in more than 20 years of experiments,” she told a packed auditorium in the “Future of Biophysics” symposium. Amaro pointed out that mutations of p53 are found in half of all human cancers, equivalent to 600,000 new patients every year. “p53 is known as the ‘guardian of the genome’,” she explained. “The mutations cause p53 to become inactive, and in those conditions cancer cells are able to proliferate.”
In some cases, though, p53 has been found to be reactivated, which in turn stops the spread of the cancer. The problem is that no-one knows what switches the p53 back on. “Small-molecule reactivation of p53 is the dream of cancer biologists,” Amaro said.
Amaro’s group has exploited simulations based on molecular dynamics to identify a number of different compounds that could reactivate p53. Computational analysis has also helped to reveal sites within biological molecules to which drug compounds are able to bind.
The success with p53 illustrates how computational and data science will play an increasingly important role in biomedical research. Amaro pointed out that both synchrotrons and electron microscopes have yielded large datasets describing many different biomolecular structures and that if these were made more widely available, they could offer exciting new opportunities for high-throughput computational studies.
Indeed, Amaro co-directs the Drug Design Data Resource, which aims to unlock pharmaceutical data vaults that could be used in computational drug discovery. “We want to open up databanks held by different industrial sources to the wider research community,” she said. “Making these data freely available will help improve computer-aided drug-design methods and accelerate the discovery of new and safer medicines.”
Trackback: Physics Viewpoint | Big data offers biomedical insights
Trackback: Blog - physicsworld.com